
The red curve represents the repeat length distribution for a randomly shuffled genome. We show an example below where we have several short repeats. We don’t care what the actual location of the repeat is.
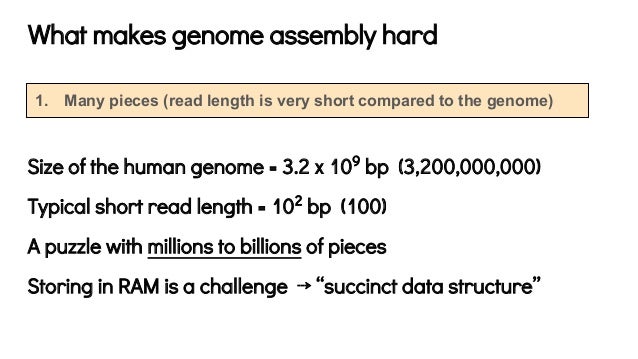
It can be seen that this bound depends only on the repeat distribution on the genome. Where \(\ell_i\) is the length of the \(i\)th repeat and the summation is over all repeats. Last time, we showed that the number of reads covering a particular location \(i\) is roughly Poisson with parameter \(c\), the coverage depth. This totals 24 distinctly-sized chromosomes.
DFIND OLD GENOME ASSEMBLIES PLUS
For example, the DNA of a human genome is organized into 22 (somatic) chromosomes numbered from 1 to 22 plus the X and Y chromosomes. Recall that we are interested in assembling a genome from individual reads. In this context, consider genome as the entire DNA (genetic material) contained within an individual cell of an organism. Therefore, many assemblies stay indefinitely as collections of contigs and/or scaffolds regardless of the organism's genomic complexity.Lecture 6: Assembly - Greedy Algorithm Lecture 6: Assembly - Greedy Algorithm Note that researchers may halt their sequencing/assembling efforts once they gather the information they need. While it is possible to generate complete assemblies for prokaryotes - where genomes are single circular chromosomes - and lower eukaryotes (such as yeast), this level is currently unattainable for the complex genomes of higher eukaryotes (including human). Finally, a non-nuclear genome (such as mitochondrion) may complete the collection for the assembly.Ĭomplete assemblies will have no sequencing gaps in the assembled chromosomes. In addition to the primary assembly, a genome assembly may contain other sequence records, such as patches (to correct regions of the primary assembly) and/or alternative loci (to offer alternative models for highly variable regions of chromosomes). Unlocalized* and unplaced** contigs and scaffolds records may accompany the chromosome records and together constitute the primary assembly. An assembly at the chromosome level will generally have one record for each chromosome. Again, researchers represent any sequencing gaps in an assembled chromosome with NNN's. The next step is to have the scaffolds that belong to the same chromosome properly ordered, oriented, and assembled into the chromosome sequence. Assemblies at the scaffold level will generally have a number of scaffold records plus a number of contigs records.
DFIND OLD GENOME ASSEMBLIES SERIES
To make a scaffold a single sequence unit (a single sequence record), they represent sequencing gaps between the contigs in the scaffold with series of NNN’s (instead of DNA sequence of A, T, G, and C). To build a scaffold, researchers place several contigs in the correct order and orientation. The researchers used blood samples from Tater, a 6-year-old Pallas’s cat who lives at the Utica Zoo in New York, to construct a high-quality diploid nuclear genome assembly, a representative map of genes for the species. The next step is to build scaffolds (supercontigs). This approach is termed Whole Genome Shotgun ( WGS) sequencing.Ĭontigs are the first level in the hierarchy of a genomic assembly. Genome assembly is the process of reconstructing a genome from randomly sampled sequence fragments in which overlapped sequence fragments are referred to as contigs (in the ideal case, one. Once the sequences of the small pieces - called reads - are obtained, researchers assemble these like tiny pieces of a giant puzzle into progressively larger contiguous sequence pieces (called contigs). A major strategy to generate an assembly involves (1) isolation of genomic DNA from a biological sample and (2) fragmentation of DNA into small pieces that are then sequenced individually.


 0 kommentar(er)
0 kommentar(er)
